Neuron Image Segmentation
This project was done for a Medical Imaging class at UVic.
Skills
- Computer Vision
- Convolutional Neural Networks (CNN’s)
- Image Segmentation
- Python, Tensorflow
Background
The goal of this project was given a dataset of photos of neuron dentrites that are photographed using pseudo 3D focusing was to segment out in focus areas of that neuron.
Two approaches were made. One using classical image processing techniques like morphological operations. The other approach used a CNN with a unet architecture.
CNN Approach
I decided to use a U-net architecture as they work really well for this type of segmentation task. The complete architecture is:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
def unet(input_shape=(512, 256, 1)):
inputs = Input(input_shape)
# Downscaling path
c1 = Conv2D(16, (3, 3), padding='same')(inputs)
c1 = BatchNormalization()(c1)
c1 = Activation('relu')(c1)
p1 = MaxPooling2D((2, 2))(c1)
c2 = Conv2D(32, (3, 3), padding='same')(p1)
c2 = BatchNormalization()(c2)
c2 = Activation('relu')(c2)
p2 = MaxPooling2D((2, 2))(c2)
# Bottom of the U
c3 = Conv2D(64, (3, 3), padding='same')(p2)
c3 = BatchNormalization()(c3)
c3 = Activation('relu')(c3)
# Upscaling path
u1 = Conv2DTranspose(32, (2, 2), strides=(2, 2), padding='same')(c3)
u1 = concatenate([u1, c2])
c4 = Conv2D(32, (3, 3), padding='same')(u1)
c4 = BatchNormalization()(c4)
c4 = Activation('relu')(c4)
u2 = Conv2DTranspose(16, (2, 2), strides=(2, 2), padding='same')(c4)
u2 = concatenate([u2, c1], axis=3)
c5 = Conv2D(16, (3, 3), padding='same')(u2)
c5 = BatchNormalization()(c5)
c5 = Activation('relu')(c5)
# Output layer
outputs = Conv2D(1, (1, 1), activation='sigmoid')(c5)
model = Model(inputs=[inputs], outputs=[outputs])
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
return model
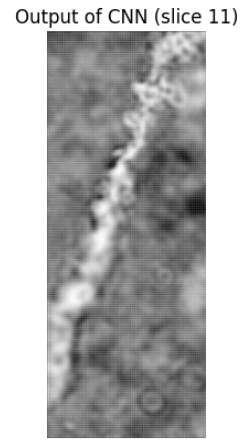
This approach did not work super well ending up with a final F1 score on the test set of ~0.4.
An example of the output for a given image is shown below:


Non ML Approach
This approach involved 5 steps:
- Homomorphic Filter
- OTSU Thresholding
- Sobel Filter
- Morphological Closing
- Combine results from step 2 with results from step 4
This ended up working better with a final F1 score of 0.6.